Calibration of genetic effect from founder population data
gen_effect_calibration.RdCalibration of genetic effect from founder population data
Arguments
- founder_object
Output of
generate_founder()orread_input_data()function- taxa_assign_g
Factor vector giving cluster assignment for all taxa, typical output of assign_taxa()
- correlation
Correlation between taxa within the same cluster, value between 0 and 1, DEFAULT = 0.5
- effect.size
Vector giving the size of genetic effect to try
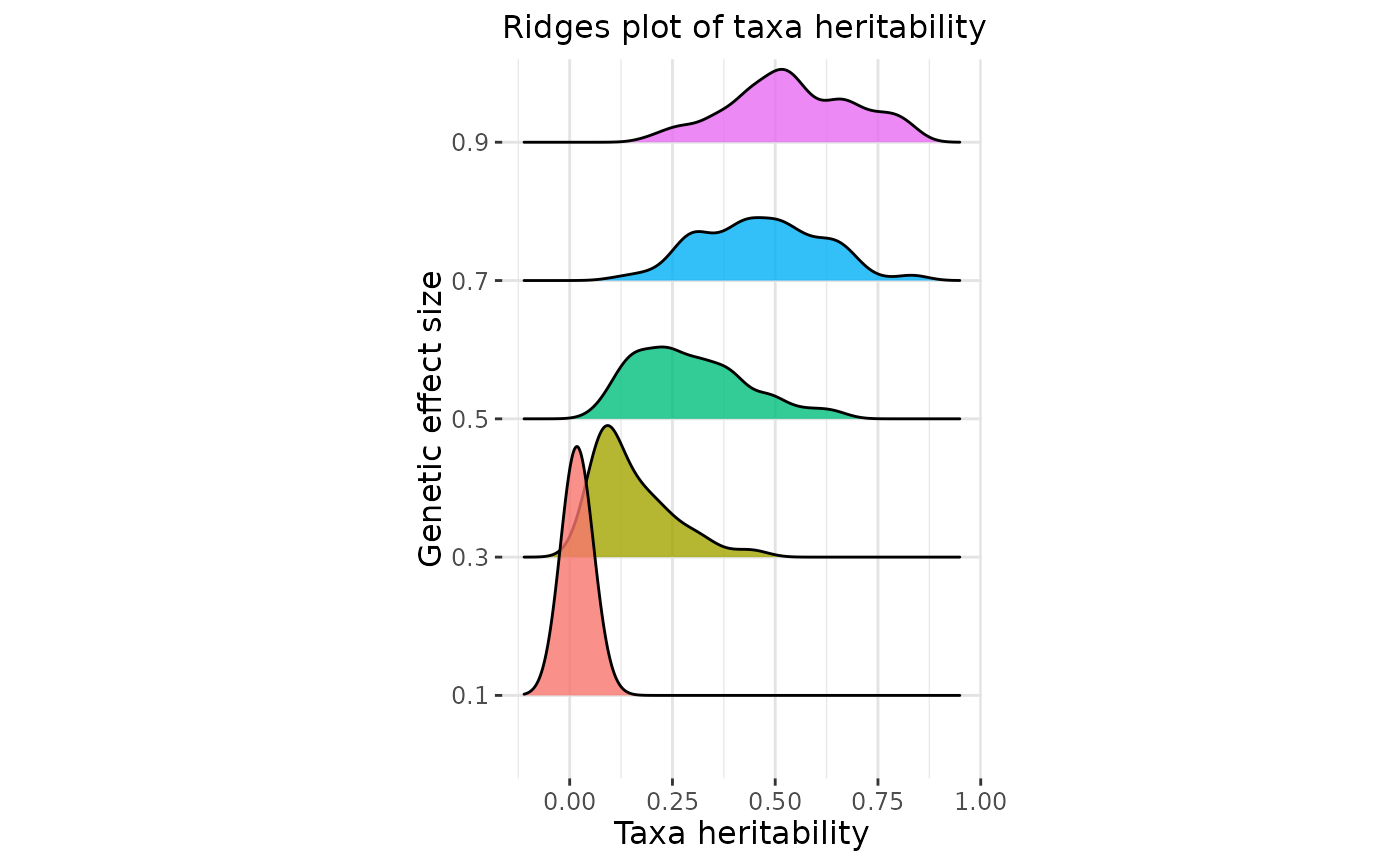
- plot
boolean, if plot generation is required, DEFAULT = TRUE
Value
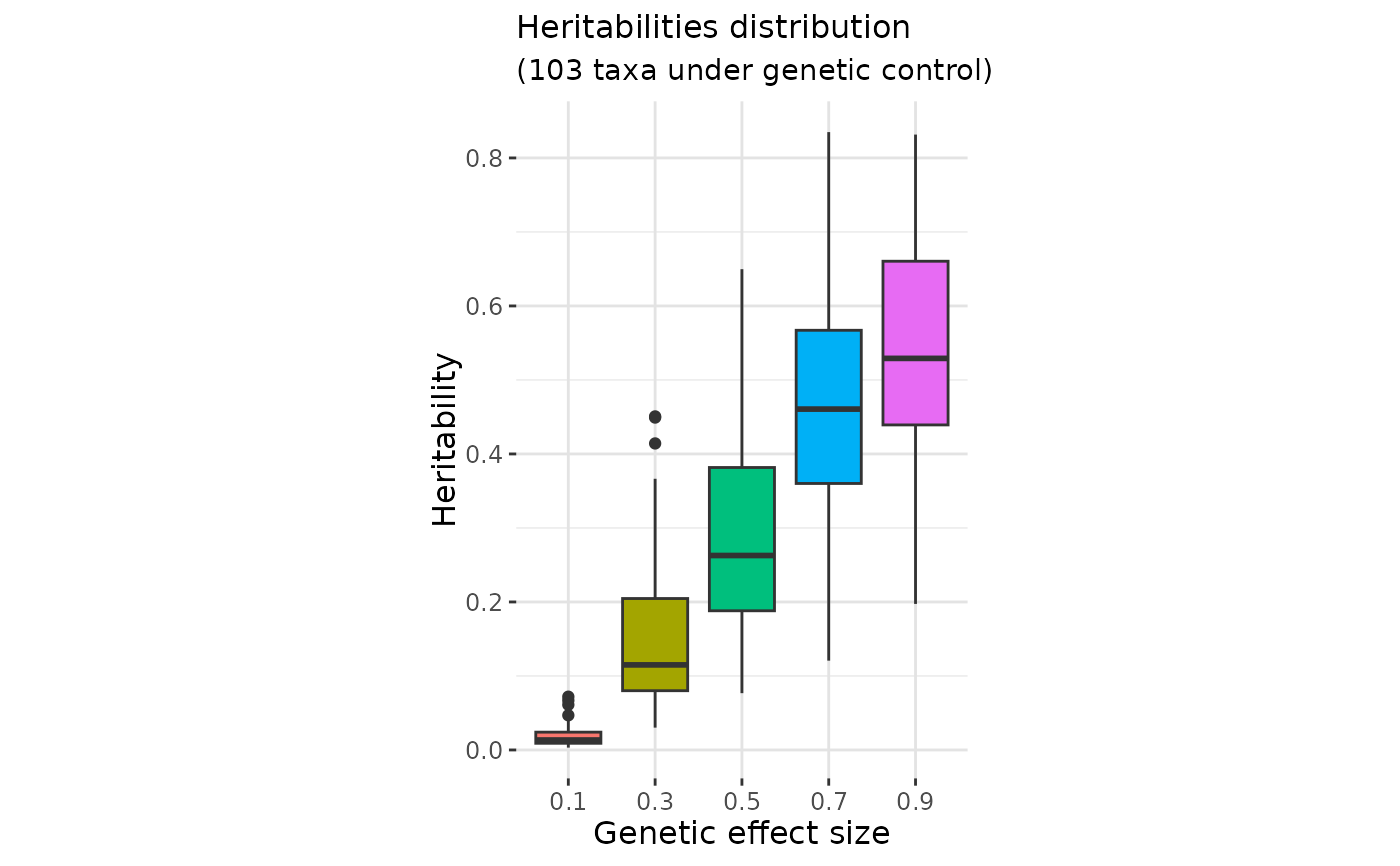
A data.frame with three columns, giving the Taxa ID, the effect.size and the corresponding heritability
Examples
data("Deru")
ToyData <- Deru
taxa_assign_g <- assign_taxa(founder_object = ToyData)
effect_size_vector <- c(seq(0.1,1, by = 0.2))
out_data <- gen_effect_calibration(founder_object = ToyData,
taxa_assign_g = taxa_assign_g,
correlation = 0.5,
effect.size = effect_size_vector,
plot = TRUE)
 #> Picking joint bandwidth of 0.0299
#> Picking joint bandwidth of 0.0299